Author: Andreas Halman, PhD on August 27, 2024 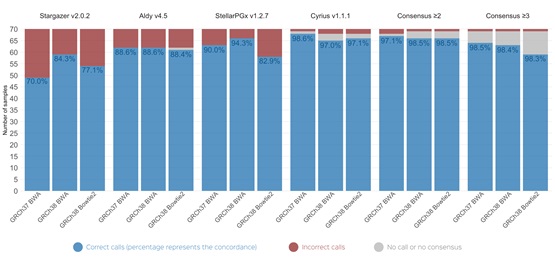
Pharmacogenomics (PGx) is a field within personalized medicine that uses genetic information to inform drug prescribing and monitoring. Several computational tools have been developed to analyze high-throughput sequencing data and genotype genes involved in drug metabolism. For many pharmacogenes, star alleles are reported, which consist of one or a combination of several genetic variants, including structural variants. In our study, we benchmarked four PGx tools and assessed their performance on six pharmacogenes, with a particular focus on CYP2D6, which is an important but challenging gene to analyze.
To gain a more comprehensive understanding, we created several datasets from the same samples, using different reference genomes, aligners, and varied sequencing depths to investigate how upstream processes impact the results of these tools. While the performance of tools on genes other than CYP2D6 was highly similar, we observed more variations in genotyping the complex CYP2D6 gene. Notably, we saw that the results from the tools could be influenced by the reference genome and sequencing aligner used. For instance, certain tools incorrectly reported the presence of a rare star allele for some samples when those were aligned on the GRCh38 reference genome and a different allele when using the older GRCh37 assembly.
Overall, our findings indicate that differences in upstream processing can, in some cases, affect the results of PGx tools and potentially influence prescribing recommendations. Therefore, using multiple high-performing tools together and adopting a consensus approach could help reduce false positives and improve the confidence of results, especially on lower sequencing depth. This is particularly important in clinical research, where accuracy is crucial for providing correct drug dosing recommendations. Read our publication to find out more about tools' performance and how reference genomes, aligners, and sequencing depth impacted the results.

The comment feature is locked by administrator.